In June 2024 I suddenly started experiencing headaches and migraines with unusual and intense pain patterns. At first, I didn’t think too much of them, but after a couple of weeks of this happening just about every day, I had decided to start seeing some doctors about it. Nothing concrete was found at first, and eventually it was recommended that I have an MRI scan.
Thankfully, the scan was normal, and I was merely diagnosed with a new presentation of normal migraines which behave quite differently than when I was younger, which is apparently a thing that just happens sometimes. It’s a bit annoying when your medical diagnosis functionally boils down to “sucks to be you,” but such is life sometimes. At least I was lucky enough to have the reassurance that everything was fine and some helpful advice on how to deal with them.
In the weeks leading up to the MRI appointment I became enamored with the principles, physics, and engineering of MRIs. They’re incredible, mind-bending inventions, and I wish I could spend some time studying them directly.
I had mentioned this whole episode to some friends in a group chat, and one of them pointed out that I could 3D print the MRI and have a 1:1 scale model of my own brain on my desk.
Well that’s obviously an opportunity I simply can’t pass up, and my 3D printer has been wanting for a new project.
I’ve seen this done before occasionally and reading up on MRI data formats weeks earlier meant I had a vague idea of how it could be done, so I had the hospital send me a copy of my MRI data on a CD. The disc arrived fairly quickly, and I immediately threw the data onto my NAS and started digging into it.
First thing’s first: Finding a way to actually read the data. This is actually really easy. It’s stored in a well-known standard format called DICOM (Digital Imaging and Communications in Medicine). Learning a bit about that is pretty neat, but I’m more interested in understanding the MRI data itself so that I can parse and convert it. I Googled around and found 3D Slicer, a visualization tool specifically designed to handle medical DICOM data. There are several others out there, but this one is entirely free, and it looked like there was enough documentation to work out how to use it. Bonus points in that there are several plugins available to accomplish this exact task of isolating the brain out of my data and representing it as a 3D model. Awesome!
Alright, so now I need to configure these plugins to automate the segmentation of my data. Right now, it’s a series of roughly two thousand 2D images which represent planar slices of my head. I’m only interested in the brain, not so much my skull or neck. You can either very tediously select the relevant object by hand, or automate this process. I decided to take the machine learning approach and use pre-trained models from MONAI and a tool called FSL to throw a neural network at the problem. This process resulted in converting the DICOM data to another format called NIfTI which contained the segmented brain imaging data. This is the stage that took up most of my time on this project, as I spent a lot of my day working through the docs and prodding at the tools.
At this stage with a hint of morbid curiosity I actually ended up asking Google Gemini to help guide me through this process; partly just to see if it could deal with something as complex as this and to see if my understanding of the documentation was on the right track. For all the discourse about LLM-based AIs (of which I have many thoughts, but they’re outside the scope of this particular blog post), they can be useful rubber ducks in a pinch as long as you’re careful and understand their limitations and pitfalls. Much to my surprise, it actually yielded rather helpful and largely correct insights on all the aforementioned tools and taught me some useful functions for them all. Once I had the relevant model running on my laptop and 3D Slicer connected to it, it was finally time to create the 3D rendering and output the STL file.
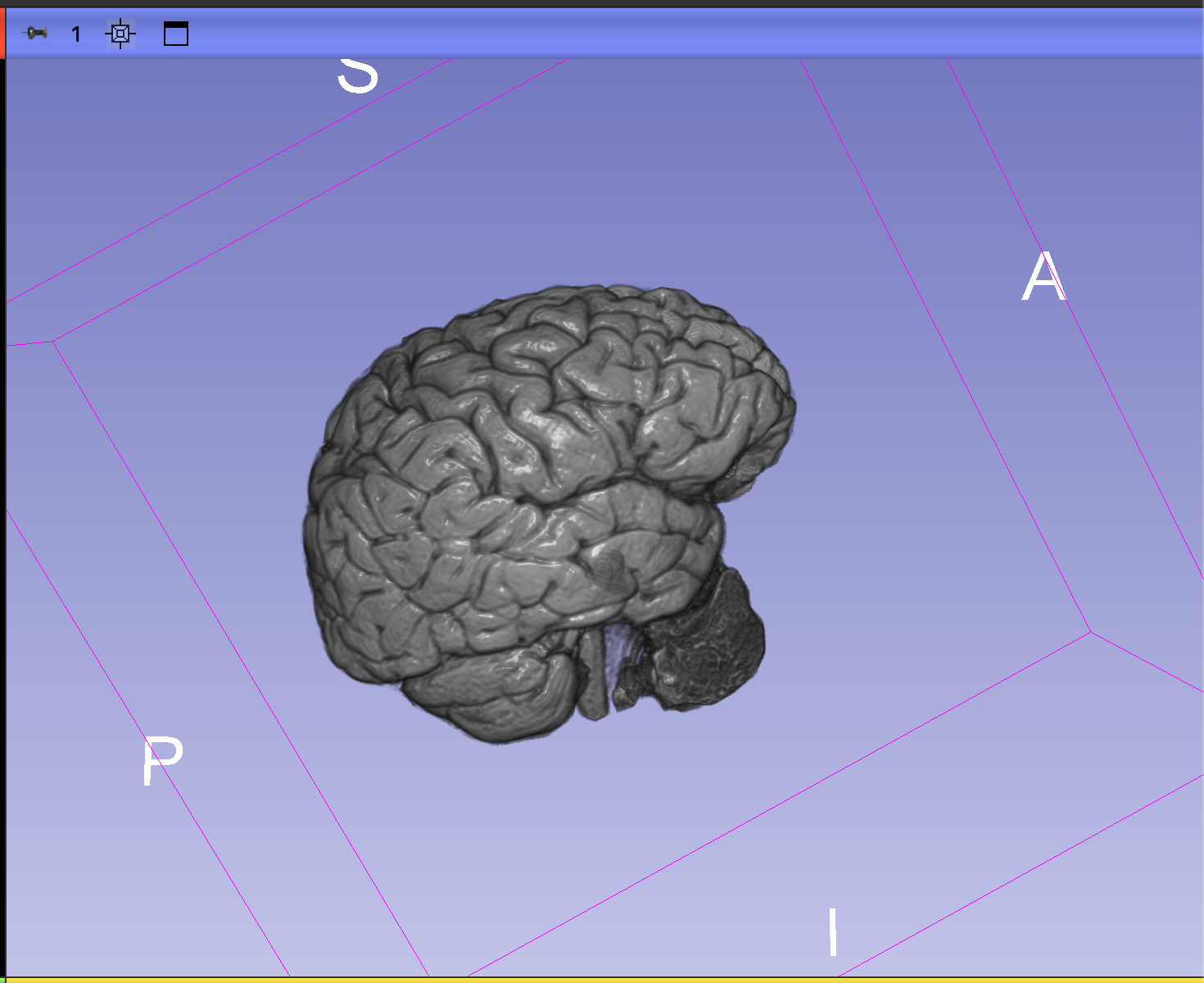
After several hours of research and subsequent trial and error, I finally managed to successfully segment my brain out of my MRI data and create a 3D printable model from it! The final version of the STL file required some finessing to bring out the topographical details and clean up some noise, but my best result was good enough as far as I’m concerned. I now have a 1:1 scale 3D model of my own brain! Pretty damn cool.
I put the model in my 3D printer slicing software, made some tweaks to the print for reliability and prepped the printer. I didn’t actually have appropriately colored filament immediately on hand, so I just used a lovely shade of blue I had lying around and decided to print the model at 40% scale as a test print. Sent that to my printer, and 12 hours later, I had myself a copy of my own brain in the palm of my hand!
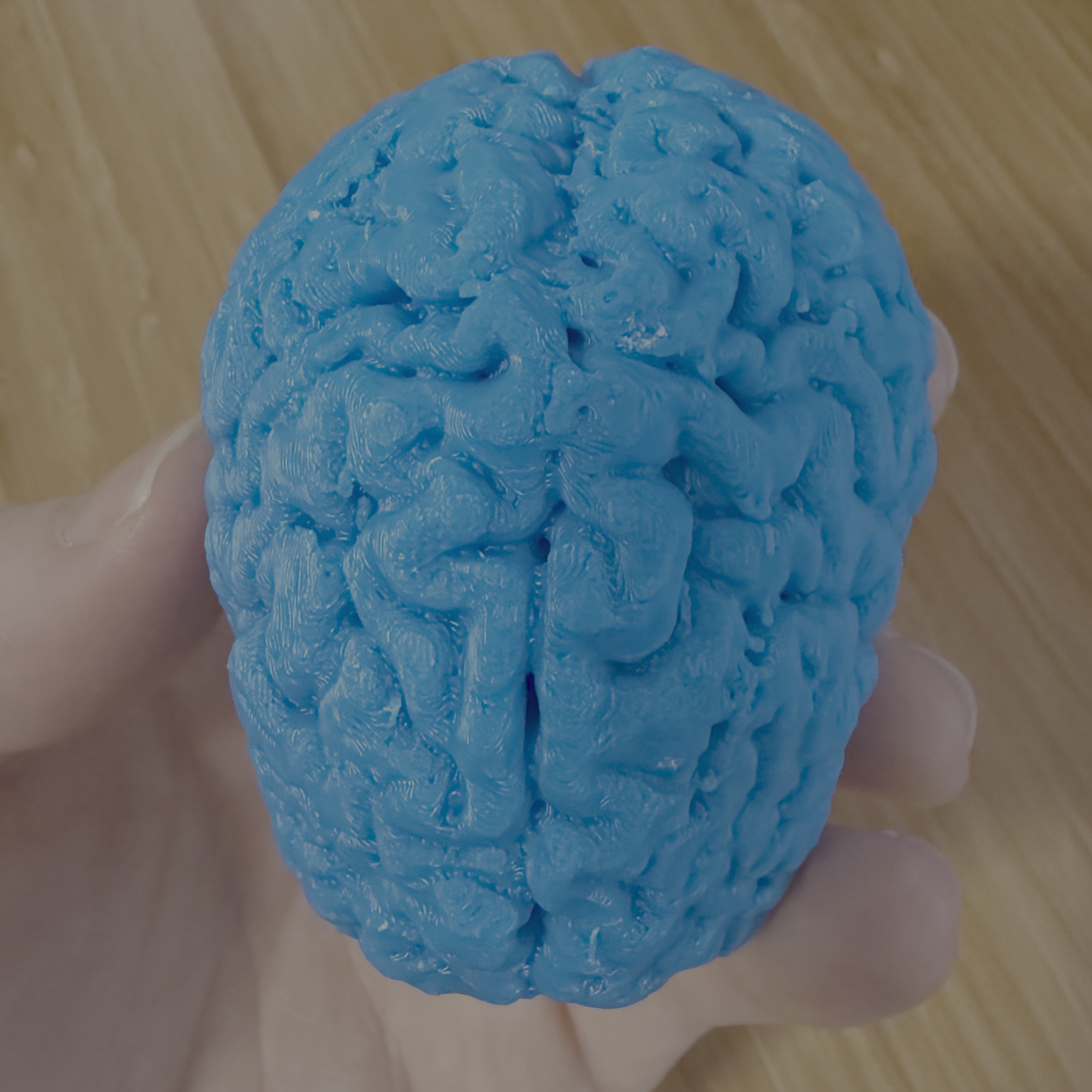
Rad.
I’ve ordered some pink filament and plan to print this model at 1:1 scale. I may have some annoying migraines these days, but at least now I have a tangible object to place the blame on.